XYZ file reader

User interface of the XYZ reader, which appears as part of a pipeline’s file source.
XYZ is a simple text-based file format for storing particles or atoms and their properties.
Basic XYZ format
In its simplest form, an XYZ file consists of a short header (two lines) followed by the list of atoms:
8
Cubic bulk silicon cell
Si 0.00000000 0.00000000 0.00000000
Si 1.36000000 1.36000000 1.36000000
Si 2.72000000 2.72000000 0.00000000
Si 4.08000000 4.08000000 1.36000000
Si 2.72000000 0.00000000 2.72000000
Si 4.08000000 1.36000000 4.08000000
Si 0.00000000 2.72000000 2.72000000
Si 1.36000000 4.08000000 4.08000000
The first line specifies the number of atoms and the second line is used to store an arbitrary comment text (may be an empty line). Each of the following atom lines consist of the species name and the atom’s Cartesian xyz coordinates.
This reader is able to load gzipped XYZ files (“.gz” suffix) and zstd compressed files (“.zst” suffix).
XYZ files may store simulation trajectories. Multiple frames are simply stored back-to-back in one file, i.e., the next two-line header directly follows after the atoms list of the preceding frame. OVITO automatically detects if the loaded XYZ file contains more than one frame.
Simulation cell and boundary conditions
Since the basic XYZ file format doesn’t contain any information about a simulation cell or boundary conditions, OVITO assumes that the XYZ file describes a non-periodic system.
The file reader option Generate bounding box if needed can be enabled to instruct OVITO to create a tight simulation cell by computing the axis-aligned bounding box of the loaded particle coordinates during import.
This ad-hoc cell is useful for visualizing the atomic positions in a non-periodic system, but it is not suitable for periodic systems, since the generated bounding box will not match the original simulation cell geometry of the simulation. In such cases, the extended XYZ file format should be used instead for data exchange with OVITO, because it stores the true simulation cell geometry and periodic boundary conditions.
XYZ files with additional columns
While the basic XYZ format consists of exactly four data columns as described above, OVITO is prepared to read XYZ files with an arbitrary number of columns containing auxiliary per-atom attributes. The file reader will display the following dialog window to let you specify the mapping of each file column to a corresponding particle property within OVITO.
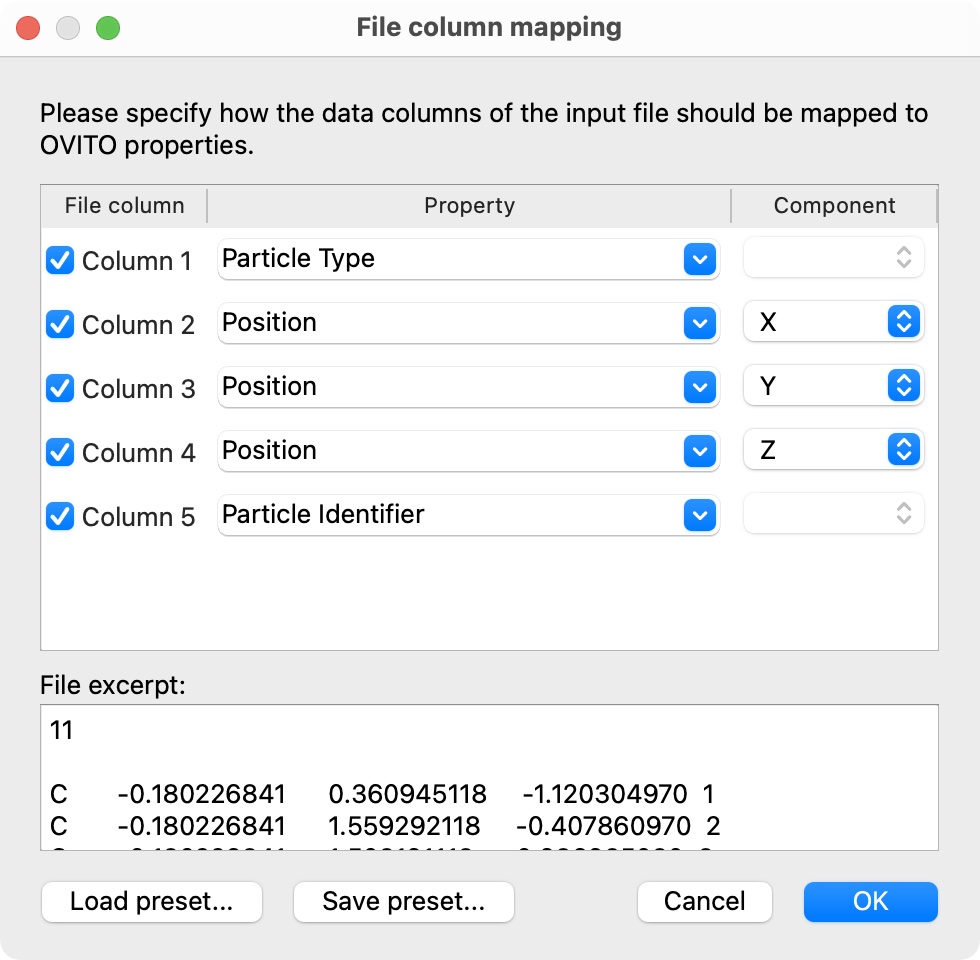
OVITO normally adopts the original order of the atoms as they are listed in the XYZ file.
However, if a file column contains unique atom IDs, and they are mapped to OVITO’s Particle Identifier
property, the file reader provides a user option to sort atoms by ID during import
(sort_particles keyword parameter in Python, see below).
Extended XYZ format
The extended XYZ format is an enhanced version of the basic XYZ format, allowing extra columns to be present in the file for additional per-atom properties as well as standardizing the format of the comment line to include the simulation cell geometry, boundary conditions, and other per-frame parameters. Here is an example:
8
Lattice="5.44 0.0 0.0 0.0 5.44 0.0 0.0 0.0 5.44" Properties=species:S:1:pos:R:3 Time=0.0
Si 0.00000000 0.00000000 0.00000000
Si 1.36000000 1.36000000 1.36000000
Si 2.72000000 2.72000000 0.00000000
Si 4.08000000 4.08000000 1.36000000
Si 2.72000000 0.00000000 2.72000000
Si 4.08000000 1.36000000 4.08000000
Si 0.00000000 2.72000000 2.72000000
Si 1.36000000 4.08000000 4.08000000
In the extended XYZ format, the comment line is replaced by a series of key/value pairs.
The keys should be strings, and values can be integers, reals, booleans (denoted by T and F for true and false) or strings.
Quotes are required if a value contains any spaces (like in the Lattice record above).
Attention
No whitespace is allowed in front of or after the equal sign. Lattice="..." is correct, Lattice = "..." is wrong.
There are two mandatory parameters that any extended XYZ file must specify: Lattice and Properties.
Other parameters - e.g. Time in the example above - can be added to the parameter line as needed
and will be imported by OVITO as global attributes.
Lattice is a Cartesian 3x3 matrix representation of the simulation cell vectors,
with each vector stored as a column and the 9 values listed in Fortran column-major order,
i.e. in the form:
Lattice="<ax> <ay> <az> <bx> <by> <bz> <cx> <cy> <cz>"
where \((a_x\ a_y\ a_z)\) are the Cartesian x-, y- and z-components of the first simulation cell vector \(\mathbf{a}\), \((b_x\ b_y\ b_z)\) those of the second simulation cell vector \(\mathbf{b}\), and \((c_x\ c_y\ c_z)\) those of the third simulation cell vector \(\mathbf{c}\). Optionally, the Cartesian coordinates of the simulation cell origin \(\mathbf{o} = (o_x\ o_y\ o_z)\) can be specified as follows:
Origin="<ox> <oy> <oz>"
Without this field, OVITO places the cell origin at the standard position (0,0,0).
The periodic boundary conditions in each cell direction may be specified as triplet of Boolean flags (F/T or 0/1), e.g.:
pbc="T T F"
If the pbc keyword is not present, OVITO assumes the simulation cell to be periodic in all three directions
(only if it’s an extended XYZ files including the Lattice keyword!).
The list of data columns in the file is described by the Properties parameter, which should take the form of a series of
colon-separated triplets giving the name, format (S for string, R for real, I for integer) and number of columns of each property.
For example:
Properties="species:S:1:pos:R:3:vel:R:3:flagged:I:1"
indicates that the first file column represents atomic species, the next three columns represent atomic positions, the next three velocities, and the last is an single integer called flagged. With this columns definition, the line
Si 4.08000000 4.08000000 1.36000000 0.00000000 0.00000000 0.00000000 1
would describe a silicon atom at position \((4.08, 4.08, 1.36)\) with zero velocity and the flagged particle property set to 1.
In the current version of OVITO, text columns (data format S) are only allowed for the atom species or the molecule type.
The file reader automatically maps file columns to the right particle properties in OVITO if their name matches one of the following standard names (case-insensitive):
XYZ column specification |
OVITO particle property |
|---|---|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
File columns having any other name will be mapped to a new user-defined particle property of the same name.
OpenBabel exyz format
OVITO supports also the .exyz format written by OpenBabel,
which contains a comment line starting with the token %PBC.
In this variant of the XYZ format, the simulation cell geometry follows behind the atoms list as a separate section.
Python parameters
The XYZ file reader accepts the following optional keyword parameters in a call to the import_file() or load() Python functions.
- import_file(location, columns=None, bounding_box=False, rescale_reduced_coords=False, sort_particles=False)
- Parameters:
columns (list[str | None] | None) – A list of OVITO particle property names, one for each data column in the xyz file. Overrides the mapping that otherwise gets set up automatically as described above. List entries may be set to
Noneto skip individual file columns during parsing.bounding_box (bool) – If set to
Trueand the imported XYZ file does not contain a simulation cell definition, the file reader will generate an ad-hocSimulationCellby computing the axis-aligned bounding box of the loaded particle coordinates.rescale_reduced_coords – If set to
True, and if the XYZ file contains the dimensions of the simulation cell, and if all atomic coordinates are either in the range \([0,1]\) or the range \([-0.5,+0.5]\), the file reader will convert the reduced coordinates to Cartesian coordinates.sort_particles (bool) – Makes the file reader reorder the loaded particles before passing them to the pipeline. Sorting is based on the values of the
Particle Identifierproperty loaded from the xyz file, if any.
Changed in version 3.10.0: New default value rescale_reduced_coords=False.
Changed in version 3.14.0: New default value bounding_box=False.